Protein Electrophoresis
The use of molecular markers for identifying species identities
At the most recent VHS survey, at the Breaks Interstate Park, there were two species of slimy salamanders previously reported to occur there, Plethodon glutinosus (the Northern Slimy Salamander) and P. kentucki (the Cumberland Plateau Salamander). The morphological characteristics separating these two species are relative. Plethodon kentucki is a bit smaller, has a lighter chin, and fewer and smaller dorsal spots than P. glutinosus. These traits can be hard to tell if you only have one salamander and not a group including both for comparison purposes.

from Dickenson County Virginia.
Can you tell for sure, which individual in the above photo is which species? James Petranka in his excellent book Salamanders of the United States and Canada says that "Identification is most reliable using electrophoretic comparisons of proteins." Richard Highton originally described the different species of slimy salamanders on the basis of protein electrophoresis. Protein electrophoresis is a technique by which various forms of the same protein are separated on the basis of their overall electrical charge. Proteins are composed of chains of amino acids. Some amino acids have a positive electrical charge, others are negative, and yet others are neutral. When all the amino acids in a protein are considered, and the positive and negative charges added together, the protein will have an overall electrical charge being either positive or negative. Different species will have a common set of proteins their cells use for normal biochemical reactions. However, slight differences in the amino acid sequence of that protein can result in different species having a slightly different form (called isozymes) of that protein with a slightly different overall electrical charge on that protein. In electrophoresis, proteins from different animals are placed in a porous gel and an electrical charge is applied to that gel. Most proteins are negatively charged and will migrate through that gel towards the positively charged anode. If one isozyme has a slightly greater negative charge, it will migrate more rapidly, and over time a greater distance, than the isozyme with a lesser negative charge.
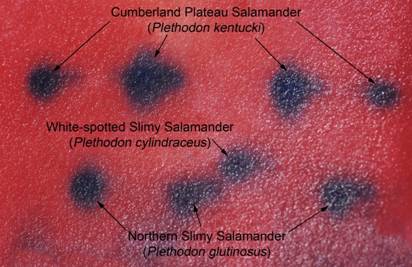
three species of slimy salamanders.
Glutamate Oxaloacetate Transaminase (GOT or AAT) is an enzyme that transfers the amino group from an amino acid to an organic acid making a different amino acid from that organic acid. It differs in amino acid sequence (and electrical charge) for three of the four slimy salamanders in Virginia, making it diagnostic for species identification. GOT for P. kentucki is the most negative in net charge and migrates the fastest and farthest, then P. cylindraceous, then the isozyme for P. glutinosus and P. chlorobryonis which is the slowest. If you have tissue samples from more than one species these can be placed in a gel, the electric field applied, and the proteins allowed to migrate through the gel for a few hours. Then the current is turned off and the gel is stained in chemicals that leave a blue-colored mark in the gel where ever the protein ended up after its migration. It is then possible to tell what species the animal was depending on how fast the protein migrated. Below in Figure 1 is a gel stained for GOT including 4 P. kentucki, 3 P. glutinosus, and one P. cylindraceous. Knowing that the isozyme for P. kentucki migrates the fastest and is at the top of the gel shows that 4 of the salamanders are P. kentucki. Knowing that the isozyme for P. cylindraceous has an intermediate migration shows there is one of this species. Since that animal was collected from an area where only that species occurs makes this a useful reference sample since it's identify is known. Finally knowing that the isozyme for P. glutinosus is slowest and closest to the bottom of the gel shows there are three of this species on the gel. No animals were included from the southeastern corner of the state so there was no possibility that P. chlorobryonis was included on the gel. If you only have one animal, it would not be possible to tell what species it was. You have to run reference samples on the same gel. However, if you have those samples, or if you know your group of animals includes both species, it may be easier to identify animals on the basis of molecular markers than with traditional morphological ones. Seeing if a protein migrates fast or slow may be easier than determining if a chin is darker or lighter, of if the spots are larger or smaller. Obviously, you need a lab and equipment to run electrophoresis, and most private individuals do not have this, but most universities have these resources. Now that I have these tissue samples identified, I have reference samples I can pull out of the freezer and use should I ever need to identify another slimy salamander.
Molecular markers have great utility for identifying different species. In this case, protein electrophoresis was used to confirm that both species of slimy salamanders were collected during the Breaks Interstate Park Survey, and to identify which animals represented which species. Proteins were used here, but DNA itself can also be used in a similar fashion to identify species, since they usually have unique genes. Forensic biologists have used DNA electrophoresis to prove that tissue samples were from protected and endangered species to shut down illegal traffic in such animals.
Paul Sattler, Ph.D.
Liberty University
Department of Biology
1971 University Blvd.
Lynchburg, VA 24501
psattler@liberty.edu



